Hyperbolic Graph Convolutional Neural Networks
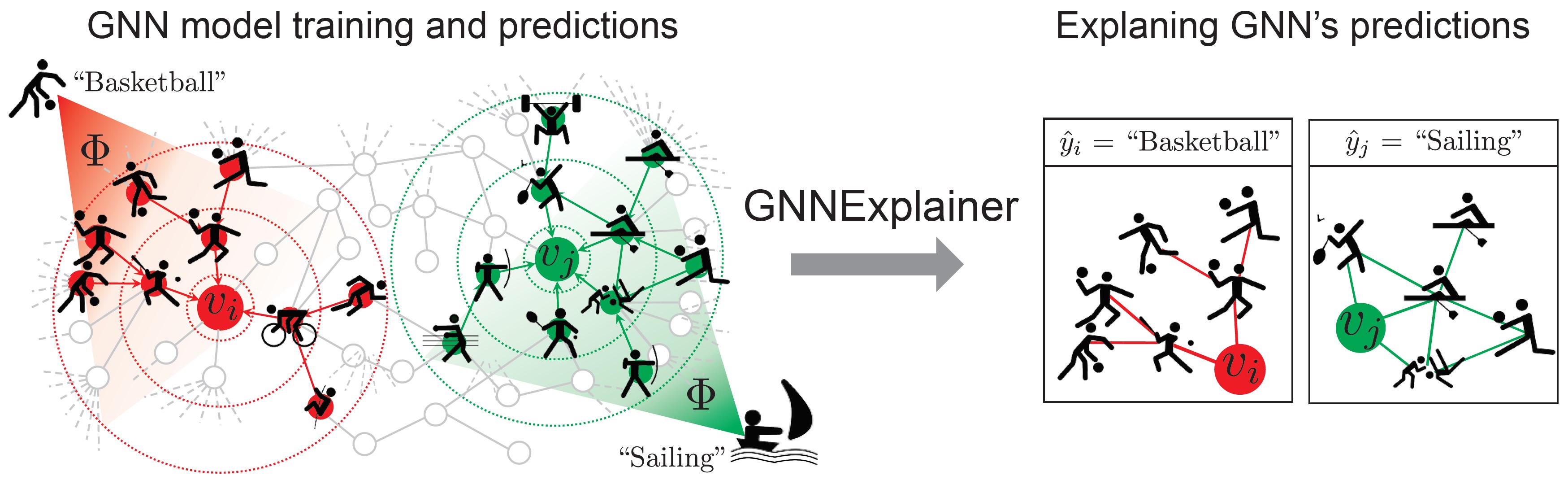
GNN-Explainer is a general tool for explaining predictions made by graph neural networks (GNNs). Given a trained GNN model and an instance as its input, the GNN-Explainer produces an explanation of the GNN model prediction via a compact subgraph structure, as well as a set of feature dimensions important for its prediction.
Motivation
Graph Neural Networks (GNNs) are a powerful tool for machine learning on graphs. GNNs combine node feature information with the graph structure by recursively passing neural messages along edges of the input graph. However, incorporating both graph structure and feature information leads to complex models and explaining predictions made by GNNs remains unsolved.
GNN-Explainer is task agnostic: it can be applied to
- Node classification
- Graph classification
- Link prediction
GNN-Explainer can be applied to many common GNN models: GCN, GraphSAGE, GAT, SGC, hypergraph convolutional networks etc.

Method
GNN-Explainer specifies an explanation as a rich subgraph of the entire graph the GNN was trained on, such that the subgraph maximizes the mutual information with GNN’s prediction(s). This is achieved by formulating a mean field variational approximation and learning a real-valued graph mask which selects the important subgraph of the GNN’s computation graph. Simultaneously, GNN-Explainer also learns a feature mask that masks out unimportant node features.
Please refer to our paper for detailed explanations and more results.Code
A reference implementation of GNN-Explainer and interactive notebook demo will be made public on Nov 15th 2019.Datasets
Mutagenicity and Reddit-Binary datasets can be found here.Contributors
The following people contributed to GNN-Explainer:
Rex Ying
Dylan Bourgeois
Jiaxuan You
Marinka Zitnik
Jure Leskovec
References
GNNExplainer: Generating Explanations for Graph Neural Networks, . R. Ying, D. Bourgeois, J. You, M. Zitnik, J. Leskovec. Neural Information Processing Systems (NeurIPS), 2019.