Disease Pathways in the Human Interactome
We perform a large-scale analysis of disease pathways in the human interactome to better understand connectivity and higher-order network structure of disease pathways.
Discovering disease pathways, which can be defined as sets of proteins associated with a given disease, is an important problem that has the potential to provide clinically actionable insights for disease diagnosis, prognosis, and treatment.
Computational methods aid the discovery by relying on protein-protein interaction (PPI) networks. They start with a few known disease-associated proteins and aim to find the rest of the pathway by exploring the PPI network around the known disease proteins. However, the success of such methods has been limited, and failure cases have not been well understood.
Here we study the PPI network structure of disease pathways. We find that 90% of pathways do not correspond to single well-connected components in the PPI network. Instead, proteins associated with a single disease tend to form many separate connected components/regions in the network. We then evaluate state-of-the-art disease pathway discovery methods and show that their performance is especially poor on diseases with disconnected pathways.
We conclude that network connectivity structure alone may not be sufficient for disease pathway discovery. However, we show that higher-order network structures, such as small subgraphs of the pathway, provide a promising direction for the development of new methods.
Publication
Large-scale analysis of disease pathways in the human interactome.
Monica Agrawal*, Marinka Zitnik* and Jure Leskovec.
Pacific Symposium on Biocomputing 2018.
Disease pathways
Disease pathways have the power to illuminate molecular mechanisms but their discovery is a challenging computational task. It involves identifying all disease-associated proteins, grouping the proteins into a pathway, and analyzing how the pathway is connected to the disease at molecular and clinical levels.
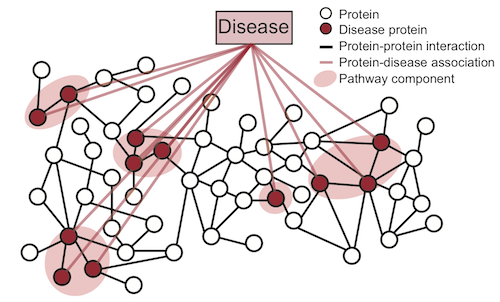
Broadly, a disease pathway in the PPI network is a system of interacting proteins whose atypical activity collectively produces some disease phenotype. In the figure below, proteins associated with a disease are projected onto the protein-protein interaction (PPI) network. Disease pathway is then a subgraph of the PPI network defined by the set of disease-associated proteins.
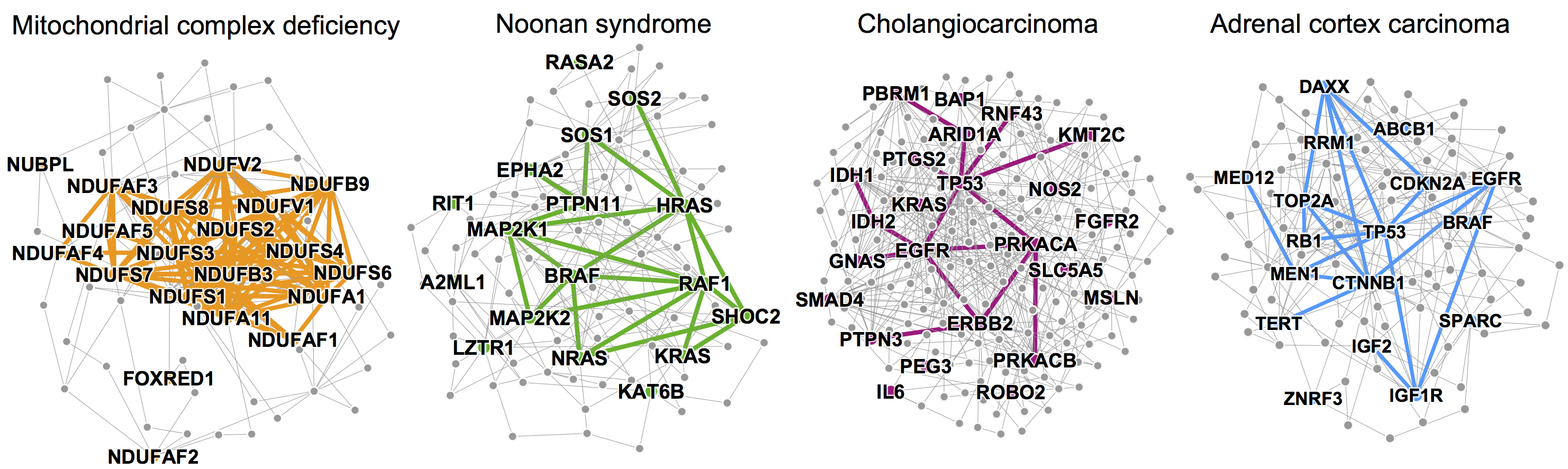
The figure below shows four disease pathways in the wider PPI network. A small PPI subnetwork highlights physical interactions between disease proteins associated with mitochondrial complex I deficiency, Noonan syndrome, cholangiocarcinoma, and adrenal cortex carcinoma.
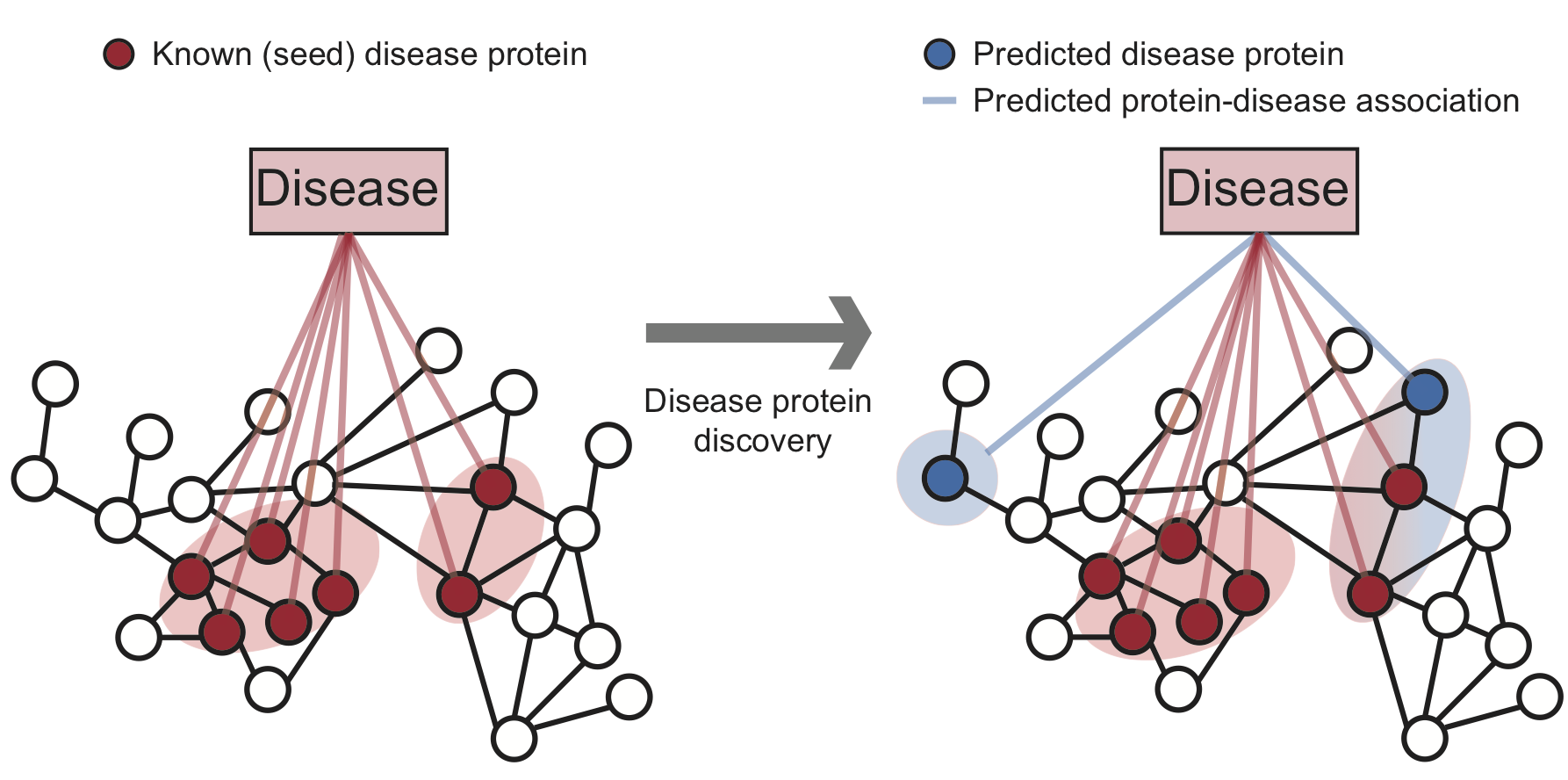
Methods for disease protein discovery predict candidate disease proteins using the PPI network and known proteins associated with a specific disease. Predicted disease proteins can be grouped into a disease pathway to study molecular disease mechanisms.
Disease pathway and protein interaction data
| File | Description |
|---|---|
| bio-pathways-network.tar.gz | Human protein-protein interaction network (nodes are gene Entrez IDs) |
| bio-pathways-associations.tar.gz | Protein-disease associations (disease pathways) |
| bio-pathways-diseaseclasses.tar.gz | Mapping of diseases to disease categories |
Supplementary tables
| File | Description |
|---|---|
| bio-pathways-features.tar.gz | Structural features of disease pathways |
| bio-pathways-diseasemotifs.tar.gz | Higher-order network analysis of disease pathways |
| bio-pathways-proteins.tar.gz | Higher-order network analysis of proteins |
Code
Source code for the analysis reported in the paper is available on GitHub.