What is SNAP?
Stanford Network Analysis Platform (SNAP) is a general purpose network analysis and graph mining library. It is written in C++ and easily scales to massive networks with hundreds of millions of nodes, and billions of edges. It efficiently manipulates large graphs, calculates structural properties, generates regular and random graphs, and supports attributes on nodes and edges. SNAP is also available through the NodeXL which is a graphical front-end that integrates network analysis into Microsoft Office and Excel.
Installation
You can go to http://snap.stanford.edu/snap/download.html to download latest version of SNAP (v1.9). Then, after unzipping, go into directory examples and open SnapExamples.sln (Visual Studio required). If your system is UNIX-based, there is also Makefile in the directory examples.
SNAP in Linux
For Linux system, you have to install g++ first. Then, take a look at the subdirectories in examples/. Copy the Makefile*.* to your folder and change the definition of variable MAIN in Makefile.ex to your program name.
Stanford has plenty of computing resources. http://itservices.stanford.edu/service/sharedcomputing/environments lists the available computing systems. We recommend corn.stanford.edu, which has 32GB of memory to handle most of the tasks.
You can simply put the source code in your home directory and use the command make.
SNAP in Windows
Under Windows system, you should install Visual Studio first (from msdn.stanford.edu). Then, open Visual Studio and create a project, add "#include YOUR_SNAP_PATH/snap/Snap.h" in your main .cpp file and include the Snap.h/cpp into your project. Also, due to the settings of SNAP, the character set must be set to Multi-byte. You can right-click on your project and go to Properties --> Configuration Properties --> General --> Projects Defaults --> Character Set --> Select "Use Multi-Byte Character Set". Then you can enjoy the powerful arsenal of SNAP!
Data Structures in SNAP
SNAP contains a STL-like library, specified in directory glib. It contains basic data structures, like vectors, hash-tables and strings. These data structures perform efficiently and supports serialization for loading and saving.
Basic Data Structures
Composite Data Structures
Containers
Saving and Loading
SNAP supports serialization for all the data structures aforementioned, as well as the network/graph structures to be introduced.
It is much more efficient to store a data structure to binary files than to text format. Saving and loading binary files for
data structures in SNAP is very simple, just via function Save and Load.
Example .cpp for basic data structure and data type in SNAP: BasicDS_DT_SNAP.cpp
Graph Manipulation in SNAP
SNAP contains a powerful graph manupulation library, specified in directory snap. It can deal with generating, manipulating as well as analyzing graphs/networks.
Graph/Network Types
- TUNGraph: undirected graph with no multi-edge
- TNGraph: directed graph with no multi-edge
- TNEGraph: directed graph with multi-edge
- TNodeNet<TNodeData>: directed graph with TNodeData object for each node
- TNodeEDatNet<TNodeData, TEdgeData>: directed graph with TNodeData on each node and TEdgeData on each edge
- TNodeEdgeNet<TNodeData, TEdgeData>: directed multi-edge graph with TNodeData on each node and TEdgeData on each edge
- If you use the T..Net class, make sure you implement saving and loading functions. Example: NodeNet.cpp
When creating a graph/network, use smart pointer (e.g. TPt<TNGraph> g) whenever possible to let the library automatically allocating/de-allocating the space for you.
Furthermore, when adding an edge, make sure the two nodes have already been added.
Playing with a Graph
We can use SNAP library to explore and analyze a graph easily.
Example:
1. Generate a G(n,m) graph (Erdos-Renyi model): Gnm.cpp
2. Get size for connected components in an undirected graph: getCC.cpp
Datasets in SNAP
SNAP contains a collection of about 50 large network datasets from tens of thousands of nodes and edges to tens of millions of nodes and edges. It includes social networks, web graphs, road networks, internet networks, citation networks, collaboration networks, and communication networks. A full list is available at http://snap.stanford.edu/data/index.html, with downloadable datasets. Some example networks include:- Social networks: online social networks, edges represent interactions between people
- Citation networks: nodes represent papers, edges represent citations
- Collaboration networks: nodes represent scientists, edges represent collaborations (co-authoring a paper)
- Amazon networks : nodes represent products and edges link commonly co-purchased products
- Twitter and Memetracker : Memetracker phrases, links and 467 million Tweets
Loading and Saving
Loading and saving network data mentioned above is similar to that of data structures. Here's an example for data as20graph.txt in the directory
example of SNAP.
Plotting and Visualization in SNAP
Plotting in SNAP
Making a plot in SNAP requires installing the software Gnuplot. Gnuplot is a plotting software with its own
grammar. Make sure that the path containing wgnuplot.exe (for Windows) or gnuplot (for Linux) is in your environmental variable $PATH.
Fortunately, SNAP contains interfaces to help you directly generate Gnuplot scripts and run Gnuplot for you. Here's a typical example of
plotting two curves with known (x,y) coordinates:
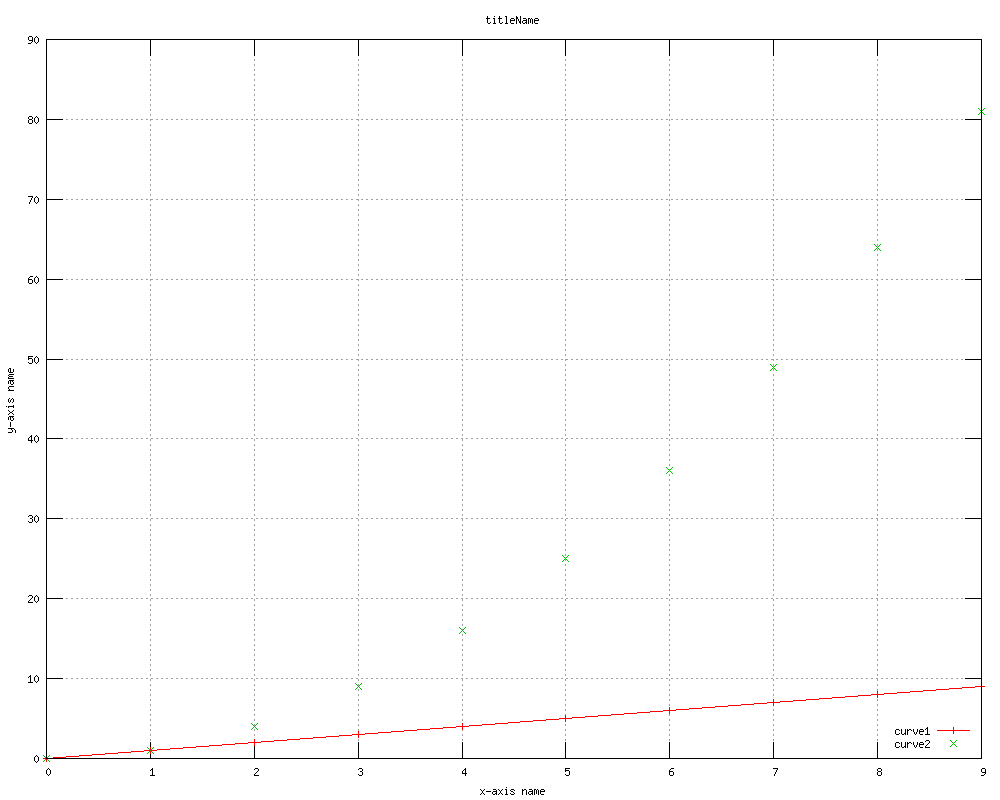 Fig. 1 Plotting line y=x and points y=x^2 using Gnuplot
Fig. 1 Plotting line y=x and points y=x^2 using Gnuplot
Typically, the plotting interface of SNAP will generate three types of files for each plot:
Example .cpp to plot exponential and Poisson distribution using Gnuplot: Gnuplot_example.cpp
Graph Visualization in SNAP
Visualize a graph in SNAP requires installing the software GraphViz. GraphViz is a graph-visualization software.
Make sure that the path of bin in your GraphViz directory is in your environmental variable $PATH. Like TGnuplot, SNAP contains interfaces to
help you directly manipulate GraphViz. Here's a typical example of visualizing a simple directed graph:
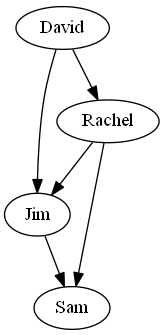 Fig. 2 Visualizing a graph in SNAP using TGraphViz
Fig. 2 Visualizing a graph in SNAP using TGraphViz