Mambo: Multimodal Biomedical Networks
Mambo is a tool for construction, representation and analysis of large multimodal networks in biomedicine.
Given a set of entities together with information about those entities, and a set of relationships between the entities together with information about those relationships, Mambo constructs a multimodal network representation of the data and provides tools for its analysis.
Recent advances in biotechnology have resulted in large and heterogeneous biomedical data generated by various experimental biotechnologies and collected in knowledge databases.
We present Mambo, a framework and a set of computational tools for construction, representation, and analysis of large-scale multimodal networks in biomedicine. Mambo scales to millions of nodes and billions of edges. Mambo supports thousands of modes (entity types), and tens of thousands of links (relationship types).
Multimodal Networks in Mambo
Mambo develops a representation of biological data that is compact and can be used to analyze large complex biomedical data and generate new domain-specific hypotheses. Multimodal networks extend the classic graph/network structure from homogeneous to heterogeneous networks. A multimodal network is composed of several set of nodes, called modes, where each mode represents a distinct entity type, connected by edges betweeen nodes within a mode and across modes.
Mambo defines a multimodal network by specifying four components:
- Nodes represent entities, e.g., the protein, kinase PKC1, and the molecular pathway, process angiogenesis.
- Modes represent node types, sets of nodes with the same semantic meaning, e.g., proteins and molecular pathways.
- Links represent edges between nodes within a mode or nodes from different modes.
- Link types represent edge types, set of edges of the same type, e.g., protein-chemical interactions and gene-disease associations.
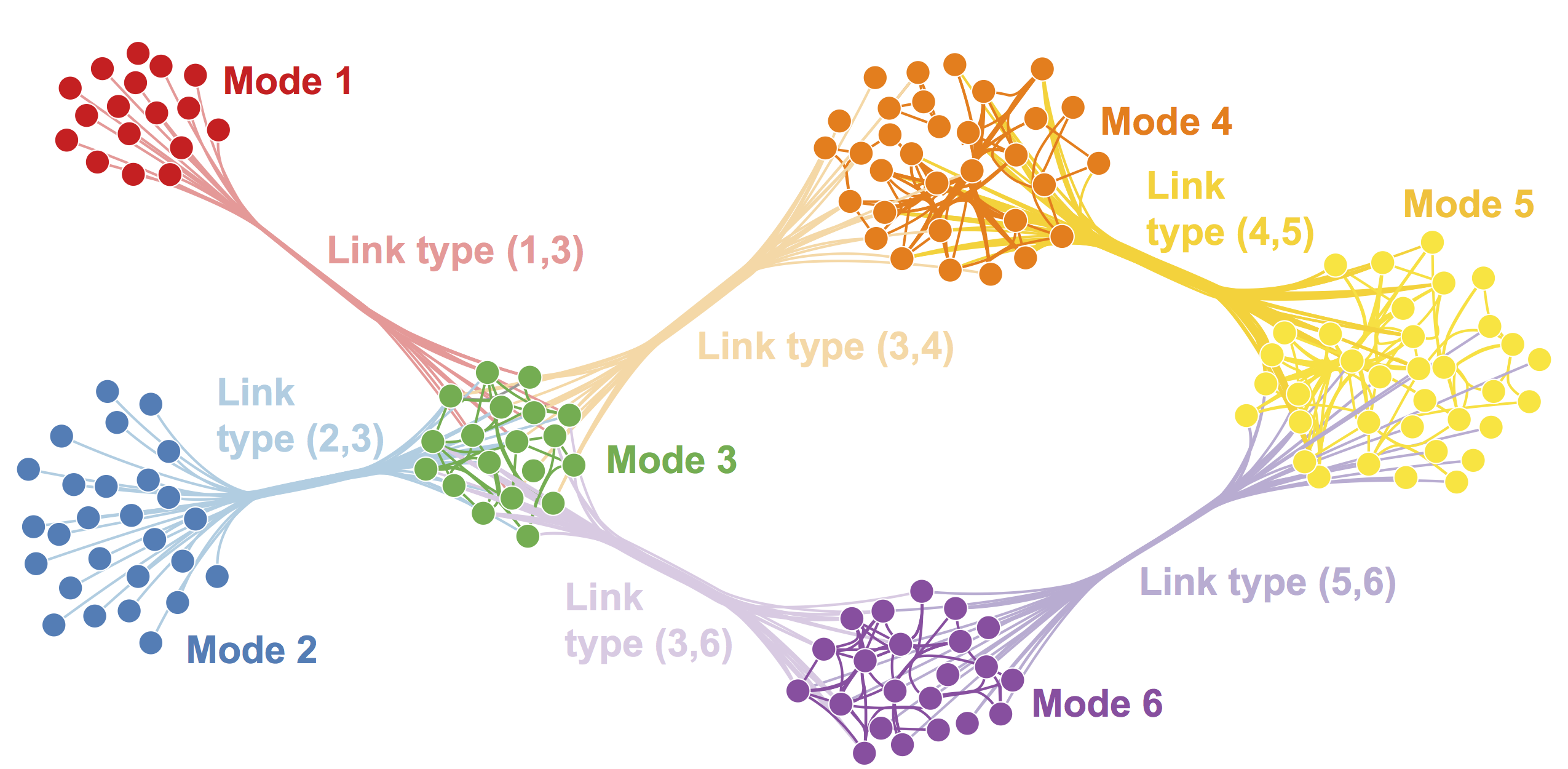
The figure below shows an example of a multimoodal network. The multimodal network consists of six modes, each denoted by a different color.
Examples of Multimodal Networks
Multimodal networks are typically constructed based a variety of data sources. We provide the processed data and the code for two multimodal networks. These examples are intended to show how to use Mambo, provide examples how to analyze the constructed networks, and to demonstrate how Mambo scales to large networks.
Multimodal Cancer Network
This network focuses on protein-coding genes that are frequently mutated in cancer patients according to the International Cancer Genome Consortium (ICGC)1. We begin with 500 most frequently mutated genes. We then include all nodes in other modes (excluding genes) that are within one-hop of these genes to construct the network based on databases listed in Data section.
We selected 500 genes with mutations in the largest number of cancer patients because these genes likely have important roles in cancer disease development and progression. The network contains information on what proteins are encoded by these genes, what drugs or chemicals interact with the genes and proteins, what functions these genes and proteins have in the human body, and what other diseases these genes and proteins are associated with.
The multimodal cancer network has 5 modes: Chemical, Disease, Function, Gene, and Protein. It has 21 link types: Chemical-Chemical, Chemical-Protein, Disease-Chemical, Disease-Disease, Disease-Function, Disease-Gene, Function-Function, Gene-Gene (split into 6 link types by interaction type), Gene-Protein, Protein-Function, and Protein-Protein (split into 6 link types by interaction type). There are a total of 20 K nodes and 3.4 M edges.
All necessary data to construct this network is in
Mambo Repository.
The repository also contains tutorial showing how to construct and analyze this network.
The tutorial begins in notebook 03 Workflow for Constructing a Multimodal Network
and ends in notebook 08 Performing Analytics on a Multimodal Network.
Giga-Scale Multimodal Biological Network
Mambo easily scales to very large datasets without any additional work required from the user. We demonstrate this property by constructing a giga-scale multimodal biological network, one of the largest networks ever constructed in biology. The network has 10 M nodes and 2.3 B edges. In this network, we integrate protein and genetic interaction data from more than two thousand species.
Further information can be found in Mambo Repository.
Setup and Installation Instructions
Prerequisites
To use Mambo and run the tutorial, please install:- Python 2.7 Download and setup instructions.
- Snap.py Downloads and setup instructions.
- Jupyter Setup instructions.
Mambo Installation
- Download notebooks from Mambo Repository.
- Launch Jupyter Notebook by clicking on the application or by issuing a command:
jupyter notebook. The web browser will open the Notebook Dashboard. Check Jupyter Notebook website for further details on how to run the notebooks. - Navigate to the directory downloaded in Step 1, start the first notebook, named
01 Introduction to Multimodal Networks, and follow the tutorial.
Mambo Tutorial
Mambo uses Jupyter Notebooks to provide a clear and easy-to-use interface with a simple set of commands that can be used to construct, represent and analyze multimodal networks. We provide a tutorial detailing how to use Mambo on several examples of real-world multimodal networks in biomedicine. Code in the tutorial can be easily adapted to new multimodal networks.
Tutorial has three parts. First, we provide background on multimodal networks and how they are represented in Mambo. This can be found in the two notebooks: 01 Introduction to Multimodal Networks and in 02 Data Representation in Mambo.
Second, we provide code and data for construction and analysis of a multimodal cancer network. This part of the tutorial provides a detailed understanding of how Mambo works. This part of the tutorial begins in notebook 03 Workflow for Constructing a Multimodal Network.
Third, we provide code for construction and analysis of a giga-scale multimodal biological network. This part of the tutorial can be found in notebook 09 Giga-Scale Multimodal Biological Network.
Detailed tutorial including background on multimodal networls, code, and data are available in Mambo Repository.
Data
The following table lists datasets used in multimodal cancer network and giga-scale multimodal biological network examples.
| Interaction Type | File | Database |
|---|---|---|
| Chemical-gene | CTD_chem_gene_ixns.tsv.gz | CTD2 |
| Disease-chemical | CTD_chemicals_diseases.tsv.gz | CTD2 |
| Disease-disease | doid.obo | Disease Ontology3 |
| Disease-function | CTD_Disease-GO_biological_process_associations.tsv.gz
CTD_Disease-GO_cellular_component_associations.tsv.gz CTD_Disease-GO_molecular_function_associations.tsv.gz |
CTD2 |
| Disease-gene | CTD_genes_diseases.tsv.gz | CTD2 |
| Drug-target | drugbank.xml.zip | DrugBank4 |
| Function-function | go.obo | Gene Ontology5 |
| Gene-gene | GeneMANIA Data | GeneMANIA6 |
| Gene-protein | ENSEMBL Data | ENSEMBL7 |
| Protein-function | goa_human.gaf.gz | Gene Ontology5 |
| Protein-protein | protein.links.detailed.v10.5.txt.gz | STRING8 |
Data References
- Zhang J. et al. International Cancer Genome Consortium Data Portal—a one-stop shop for cancer genomics data. Database, Volume 2011, 16 September 2016, bar026.
- Curated chemical–gene interactions, chemical–disease, gene–disease, disease-function data were retrieved from the Comparative Toxicogenomics Database (CTD), MDI Biological Laboratory, Salisbury Cove, Maine, and NC State University, Raleigh, North Carolina. http://ctdbase.org/.
- Kibbe W. et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Research, Volume 43, Issue D1, 28 January 2015, Pages D1071–D1078.
- Law V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Research, Volume 42, Issue D1, 1 January 2014, Pages D1091-D1097.
- The Gene Ontology Consortium. Gene Ontology Consortium: going forward. (2015) Nucleic Acids Research, Volume 43, Issue D1, 28 January 2015, Pages D1049–D1056.
- Warde-Farley D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research, Volume 38, Issue W1, 1 July 2010, Pages W214–W220.
- Bronwen L. Aken et al. The Ensembl gene annotation system. Database, Volume 2016, 1 January 2016, baw093.
- Szklarczyk D. et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research, Volume 43, Issue D1, 28 January 2015, Pages D447–D452.
Contributors
The following people contributed to the Mambo project (appear in alphabetical order):
Jure LeskovecPriyanka Nigam
Rok Sosic
Marinka Zitnik