Neural Subgraph Matching
NeuroMatch is a graph neural network (GNN) architecture for efficient subgraph matching. Given a large target graph and a smaller query graph , NeuroMatch identifies the neighborhood of the target graph that contains the query graph as a subgraph. NeuroMatch uses a GNN to learn powerful graph embeddings in an order embedding space which reflects the structure of subgraph relationship properties (transitivity, antisymmetry and non-trivial intersection), facilitating realtime approximate subgraph matching on a scale not previously possible: it can match a 100 node query graphs in a target graph of size 500k.
Motivation
Subgraph isomorphism matching is one of the fundamental NP-complete problems in theoretical computer science, and applications arise in almost any situation where network modeling is done.
- In social science, subgraph analysis has played an important role in the analysis of network effects in social networks.
- Information retreival systems use subgraph structures in knowledge graphs for semantic summarization, analogy reasoning, and relationship prediction.
- In chemistry, subgraph matching is a robust and accurate method for determining similarity between chemical compounds.
- In biology, subgraph matching is of central importance in the analysis of protein-protein interaction networks, where identifying and predicting functional motifs is a primary tool for understanding biological mechanisms such as those underlying disease, aging, and medicine.
An efficient, accurate model for the subgraph matching problem could drive research in all of these domains, by providing insights into important substructures of these networks, which have traditionally been limited by either the quality of their approximate algorithms or the runtime of exact algorithms.
Method
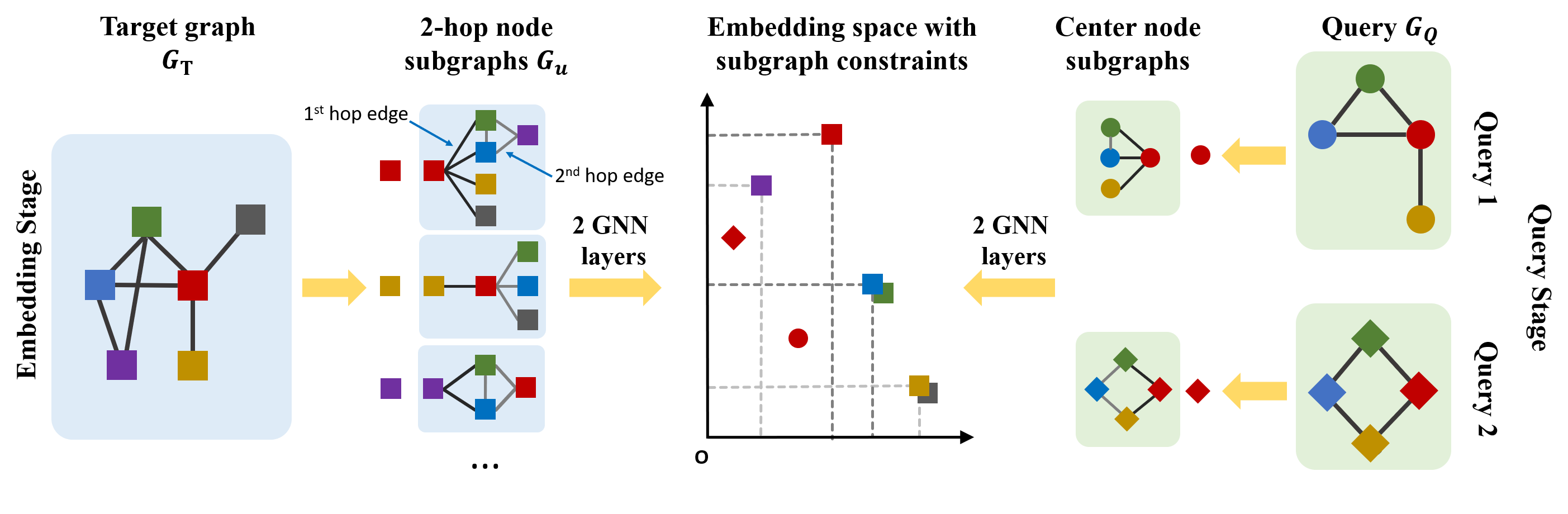
NeuroMatch treats the subgraph matching problem as a node identification problem: find node(s) in the target graph whose k-hop neighborhood contains the query. We separate this process has two parts: learning embeddings for the query and each target node, and then determining matches given the embeddings.
(1) As seen from the below figure, embeddings are learned via a GNN architecture, with one GNN applied to the target nodes (left) and one to the query nodes (right). (2) Given embeddings, we then use a simple rule for deciding whether a node is part of the subgraph: order embeddings . If every component of the embedding for the query is less than the corresponding component of the embedding for the target node, then we classify the target node's neighborhood as containing the query. We demonstrate the use of max margin loss to train the model in order to satisfy this order embedding constraint.
NeuroMatch features the following key techniques:
- Use of order embeddings constraints to improve accuracy and runtime performance.
- Optimization using node-anchored training objective, instead of learning graph-level embeddings.
- Pre-training subgraph matching on synthetic datasets and test / finetune on real datasets.
- Curriculum learning by feeding in more and more complex queries during training time.
Runtime Efficiency. Oftentimes, we have a fixed set of target graphs that we want to run queries on. In this setting, NeuroMatch excels because the embeddings for nodes in the target graph can be precomputed and stored. As queries arrive, we need only run the embedding model on the small query graph and then compare the query embedding with the stored target embeddings to perform subgraph isomorphism matching. This facilitates inference time 100x faster than previous approaches, and 10x faster than the alternative to use a learned MLP to predict subgraph relationships.
Please refer to our paper for detailed explanations and more results.Code
A reference implementation of NeuroMatch is on Github (https://github.com/snap-stanford/neural-subgraph-learning-GNN). Our code uses the DeepSNAP framework to enable efficient graph manipulation (subgraph extraction, perturbation and matching). We acknowledge Andrew Wang (awang@cs.stanford.edu) for partly helping with integration of NeuroMatch with the followup work in subgraph mining. The Repository Subgraph Learning is a general library to perform subgraph matching, and subgraph mining tasks, and can be extended to perform multiple tasks related to subgraph predictions (e.g. counting subgraphs).Datasets
Datasets for experiments can be downloaded from here. We also generate positive and negative pairs of graphs with / without subgraph relationships by sampling from the real-world and synthetic datasets. In the repository, the code for sampling can be found in common/data.py; the code for generating synthetic dataset can be found in common/combined_syn.py.Contributors
The following people contributed to NeuroMatch:
Rex Ying
Joe Lou
Jiaxuan You
Chengtao Wen, Arquimedes Canedo from Siemens
Jure Leskovec
References
Neural Subgraph Matching, . R. Ying, Z. Lou, J. You, C. Wen, A. Canedo, J. Leskovec.